12. Backpropagation
Gradient Descent Solution
def gradient_descent_update(x, gradx, learning_rate):
"""
Performs a gradient descent update.
"""
x = x - learning_rate * gradx
# Return the new value for x
return xWe adjust the old x pushing it in the opposite direction of gradx with the force learning_rate. Subtracting learning_rate * gradx. Remember the gradient is initially in the direction of steepest ascent so subtracting learning_rate * gradx from x turns it into steepest descent. You can make sure of this yourself by replacing the subtraction with an addition.
The Gradient & Backpropagation
Now that we know how to update our weights and biases using the gradient, we need to figure out how to calculate the gradients for all of our nodes. For each node, we'll want to change the values based on the gradient of the cost with respect to the value of that node. In this way, the gradient descent updates we make will eventually converge to the minimum of the cost.
Let's consider a network with a linear node l_1, a sigmoid node s, and another linear node l_2, followed by an MSE node to calculate the cost, C.
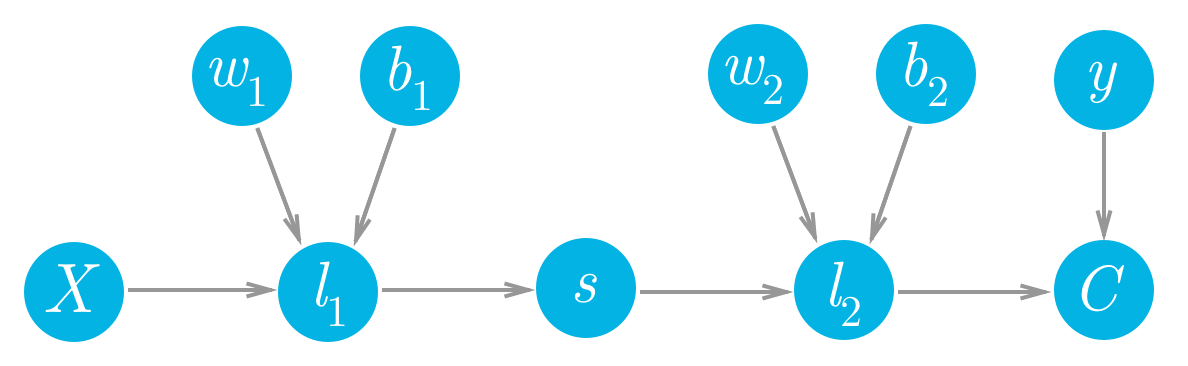
Forward pass for a simple two layer network.
Writing this out in MiniFlow, it would look like:
X, y = Input(), Input()
W1, b1 = Input(), Input()
W2, b2 = Input(), Input()
l1 = Linear(X, W1, b1)
s = Sigmoid(l1)
l2 = Linear(s, W2, b2)
cost = MSE(l2, y)We can see that each of the values of these nodes flows forwards and eventually produces the cost C. For example, the value of the second linear node l_2 goes into the cost node and determines the value of that node. Accordingly, a change in l_2 will produce a change in C. We can write this relationship between the changes as a gradient,
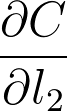
This is what a gradient means, it's a slope, how much you change the cost \partial C given a change in l_2, \partial l_2. So a node with a larger gradient with respect to the cost is going to contribute a larger change to the cost. In this way, we can assign blame for the cost to each node. The larger the gradient for a node, the more blame it gets for the final cost. And the more blame a node has, the more we'll update it in the gradient descent step.
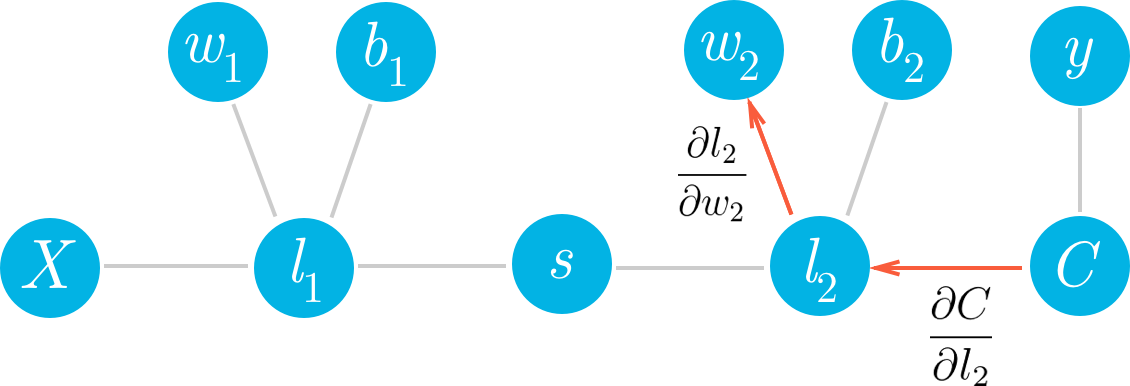
If we want to update one of the weights with gradient descent, we'll need the gradient of the cost with respect to those weights. Let's see how we can use this framework to find the gradient for the weights in the second layer, w_2. We want to calculate the gradient of C with respect to w_2:
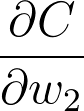
We can see in the graph that w_2 is connected to l_2, so a change in w_2 is going to create a change in l_2 which then creates a change in C. We can assign blame to w_2 by sending the cost gradient back through the network. First you have how much l_2 affected C, then how much w_2 affected l_2. Multiplying these gradients together gets you the total blame attributed to w_2.
Pre-requisites
Below we're getting into the math behind backpropagation which requires multivariable calculus. If you need a refresher, I highly recommend checking out
- Khan Academy's lessons on partial derivatives
- Another video on gradients
- And finally, using the chain rule
Continuing on
Multiplying these gradients is just an application of the chain rule:

You can see in the graph w_2, l_2, and C are chained together. Any change in w_2 will create a change in l_2 and the size of that change is given by the gradient \partial l_2 / \partial w_2. Now, since l_2 is changing this will cause a change in the cost C and the size of that change is given by the gradient \partial C / \partial l_2. You can think of the chain rule similarly to the domino effect, changing something in the network will propagate through it altering other nodes along the way.
If you think of the chain rule as normal fractions, you can see that \partial l_2 in the denominator and numerator cancel out and you get back \partial C / \partial w_2 (although it doesn't exactly work like normal fractions, but it helps to keep track of things.) Okay, let's work out the gradient for w_2. First, we need to know the gradient for l_2.
As a reminder the cost is
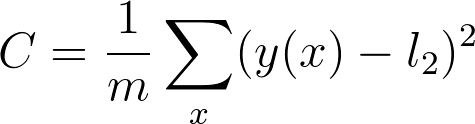
And the value for the second linear node is
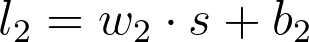
where w_2, s , and b_2 are all vectors and w_2 \cdot s means the dot product of w_2 and s .
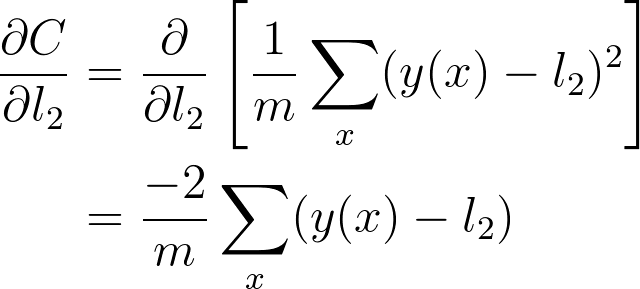
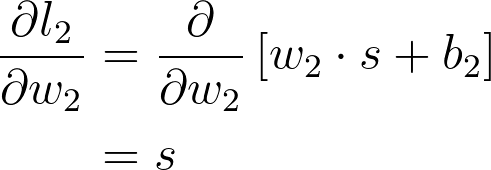
And putting these together, you get the gradient for w_2
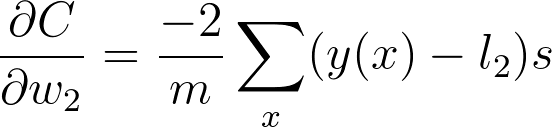
This is the gradient you use in the gradient descent update for w_2. You can see what we did here, we walked back through the graph and multiplied all the gradients we found along the way.
Now, let's go deeper and calculate the gradient for w_1. Here we use the same method as before, walking backwards through the graph.
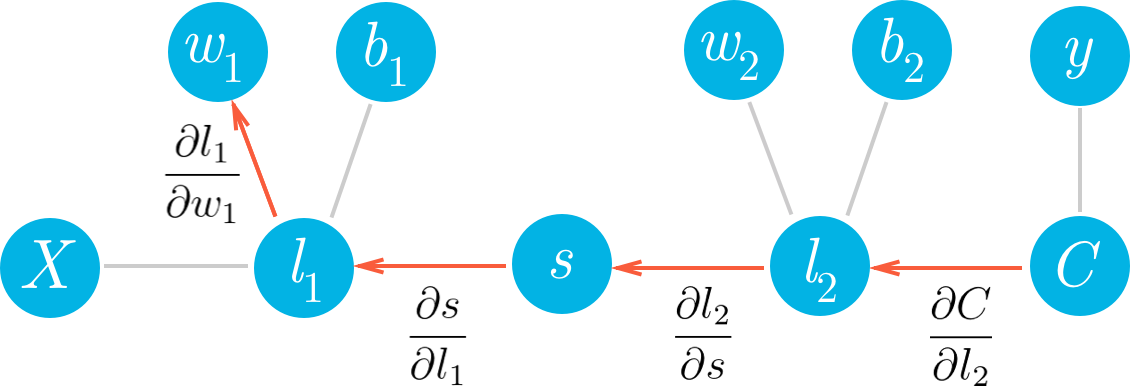
Hopefully it's clear now how to write out the gradient for w_1 just by looking at the graph. Using the chain rule, we'll write out the gradients for each node going backwards through the graph until we get to w_1.

Now we can start calculating each gradient in this expression to get the gradient for w_1
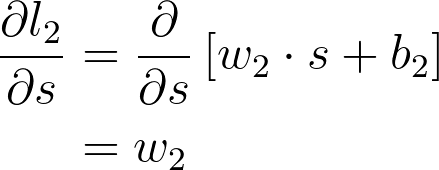
The next part is the gradient of the sigmoid function, s = f(l_1). Since we're using the logistic function here, the derivative can be written in terms of the sigmoid itself
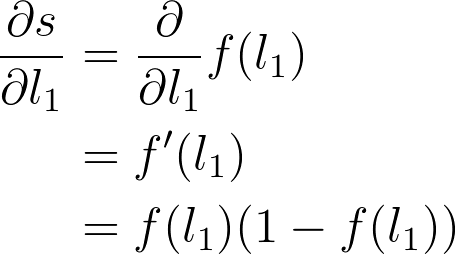

Putting this all together, you get
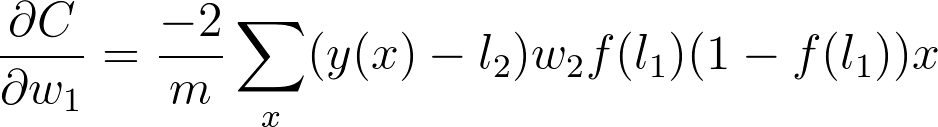
Now we can see a clear pattern. To find the gradient, you just multiply the gradients for all nodes in front of it going backwards from the cost. This is the idea behind backpropagation. The gradients are passed backwards through the network and used with gradient descent to update the weights and biases. If a node has multiple outgoing nodes, you just sum up the gradients from each node.
Implementing in MiniFlow
Let's think about how to translate this into MiniFlow. Looking at the graph, you see that each node gets the cost gradient from it's outbound nodes. For example, the node l_1 gets \partial C / \partial l_1 through the sigmoid node, s. Then l_1 passes on the cost gradient to the weight node w_1, but multiplied by \partial l_1 / \partial w_1, the gradient of l_1 with respect to it's input w_1.
So, each node will pass on the cost gradient to its inbound nodes and each node will get the cost gradient from it's outbound nodes. Then, for each node we'll need to calculate a gradient that's the cost gradient times the gradient of that node with respect to its inputs. Below I've written out this process for a Linear node.
# Initialize a partial for each of the inbound_nodes.
self.gradients = {n: np.zeros_like(n.value) for n in self.inbound_nodes}
# Cycle through the outputs. The gradient will change depending
# on each output, so the gradients are summed over all outputs.
for n in self.outbound_nodes:
# Get the partial of the cost with respect to this node.
grad_cost = n.gradients[self]
# Set the partial of the loss with respect to this node's inputs.
self.gradients[self.inbound_nodes[0]] += np.dot(grad_cost, self.inbound_nodes[1].value.T)
# Set the partial of the loss with respect to this node's weights.
self.gradients[self.inbound_nodes[1]] += np.dot(self.inbound_nodes[0].value.T, grad_cost)
# Set the partial of the loss with respect to this node's bias.
self.gradients[self.inbound_nodes[2]] += np.sum(grad_cost, axis=0, keepdims=False)New Code
There have been a couple of changes to MiniFlow since we last took it for a spin:
The first being the Node class now has a backward method, as well as a new attribute self.gradients, which is used to store and cache gradients during the backward pass.
class Node(object):
"""
Base class for nodes in the network.
Arguments:
`inbound_nodes`: A list of nodes with edges into this node.
"""
def __init__(self, inbound_nodes=[]):
"""
Node's constructor (runs when the object is instantiated). Sets
properties that all nodes need.
"""
# A list of nodes with edges into this node.
self.inbound_nodes = inbound_nodes
# The eventual value of this node. Set by running
# the forward() method.
self.value = None
# A list of nodes that this node outputs to.
self.outbound_nodes = []
# New property! Keys are the inputs to this node and
# their values are the partials of this node with
# respect to that input.
self.gradients = {}
# Sets this node as an outbound node for all of
# this node's inputs.
for node in inbound_nodes:
node.outbound_nodes.append(self)
def forward(self):
"""
Every node that uses this class as a base class will
need to define its own `forward` method.
"""
raise NotImplementedError
def backward(self):
"""
Every node that uses this class as a base class will
need to define its own `backward` method.
"""
raise NotImplementedErrorThe second change is to the helper function forward_pass(). That function has been replaced with forward_and_backward().
def forward_and_backward(graph):
"""
Performs a forward pass and a backward pass through a list of sorted nodes.
Arguments:
`graph`: The result of calling `topological_sort`.
"""
# Forward pass
for n in graph:
n.forward()
# Backward pass
# see: https://docs.python.org/2.3/whatsnew/section-slices.html
for n in graph[::-1]:
n.backward()Setup
Here's the derivative of the sigmoid function w.r.t x:
sigmoid(x) = 1 / (1 + exp(-x))
\frac {\partial sigmoid}{\partial x} = sigmoid(x) * (1 - sigmoid(x))
- Complete the implementation of backpropagation for the
Sigmoidnode by finishing thebackwardmethod inminiflow.py. - The
backwardmethods for all other nodes have already been implemented. Taking a look at them might be helpful.
Question:
Start Quiz: